Interests • Functional Basis of Demographic Trade-offs • Seed Dispersal & Seedling Regeneration • Plant-Soil Feedbacks & Soil Microbiome • Biodiveristy & Change in Nebraska's Forests
Research: Plant-Soil Feedbacks & the Soil Microbiome
Soil and root-associated microbial diversity is vast, and we lack even basic understanding of how this diversity is distributed ecologically. Using ever-evolving DNA sequencing methods, we are quantifying the structure of belowground microbial (bacteria, archaea, fungi) communities in ecosystems ranging from grasslands to tropical forests and relating their function to the plants with which they interact.
Covariation in community structure of soil bacteria and trees in Bornean rain forest
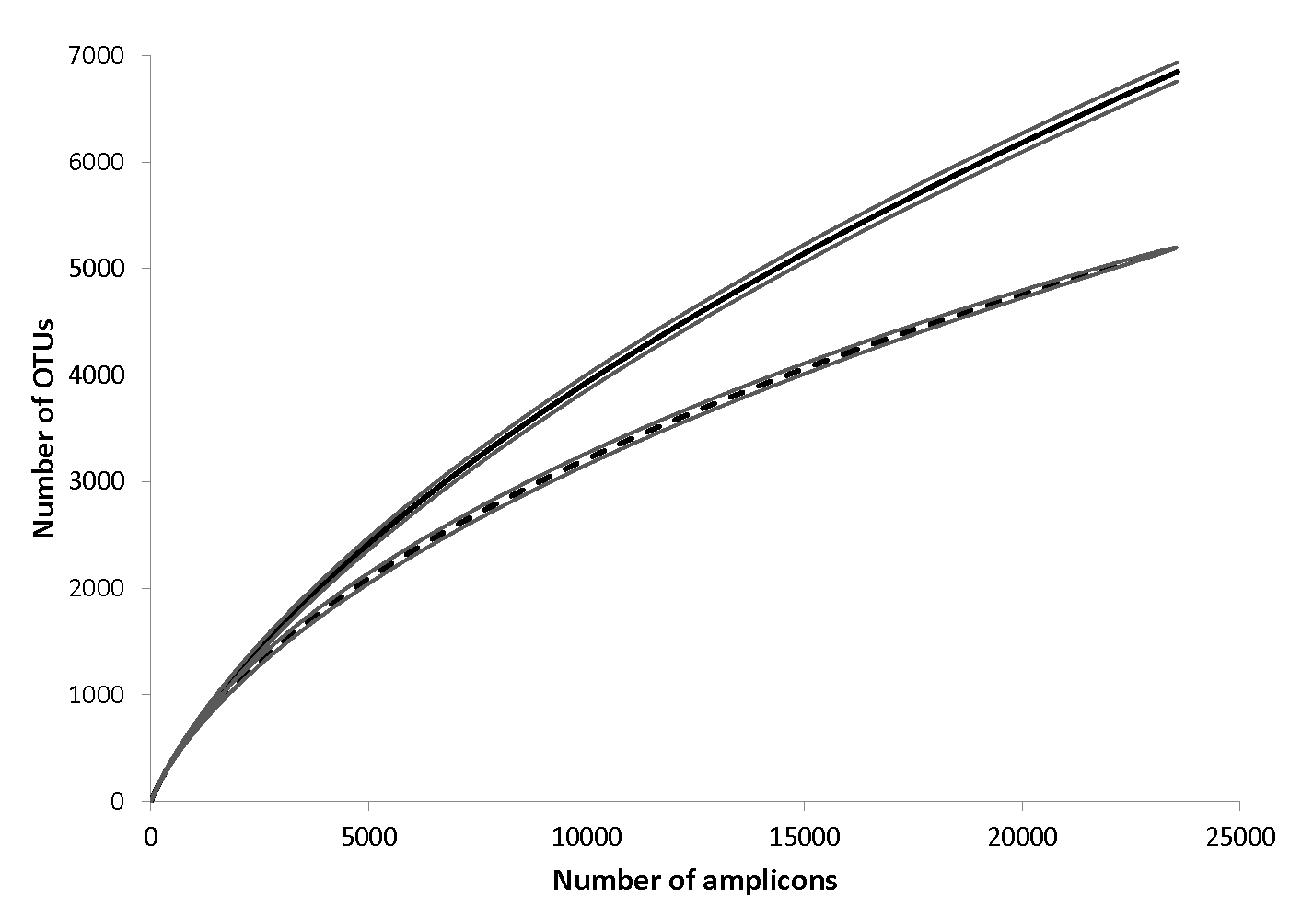
In some of the earliest work in this area, we quantified bacterial communities in two contrasting soils underlying Bornean rain forest (clay and sandy loam) that differ markedly in soil properties, aboveground tree flora, and leaf litter decomposition rates. We found significant soil-related taxonomic and phylogenetic differences between communities that, due to their proximity, are independent of climate. Bacterial communities showed distinct compositional and taxon abundance distributions that were significantly correlated with the structure of the overlying tree community. Richness of bacteria was greater in the more resource-rich clay soil. Phylogenetic community analyses suggested that environmental filtering may be an important mechanism of community assembly in clay, compared to niche-competition in sandy loam. Our study, which is the first to quantify edaphic variation in bacterial communities in soils underlying one of the most tree species rich forests on Earth, indicates an important role of plant-soil feedbacks linking the community structure of the trees and soil microbiome. (Russo et al. 2012).
Role of mycorrhizal fungi in structuring Bornean rain forests
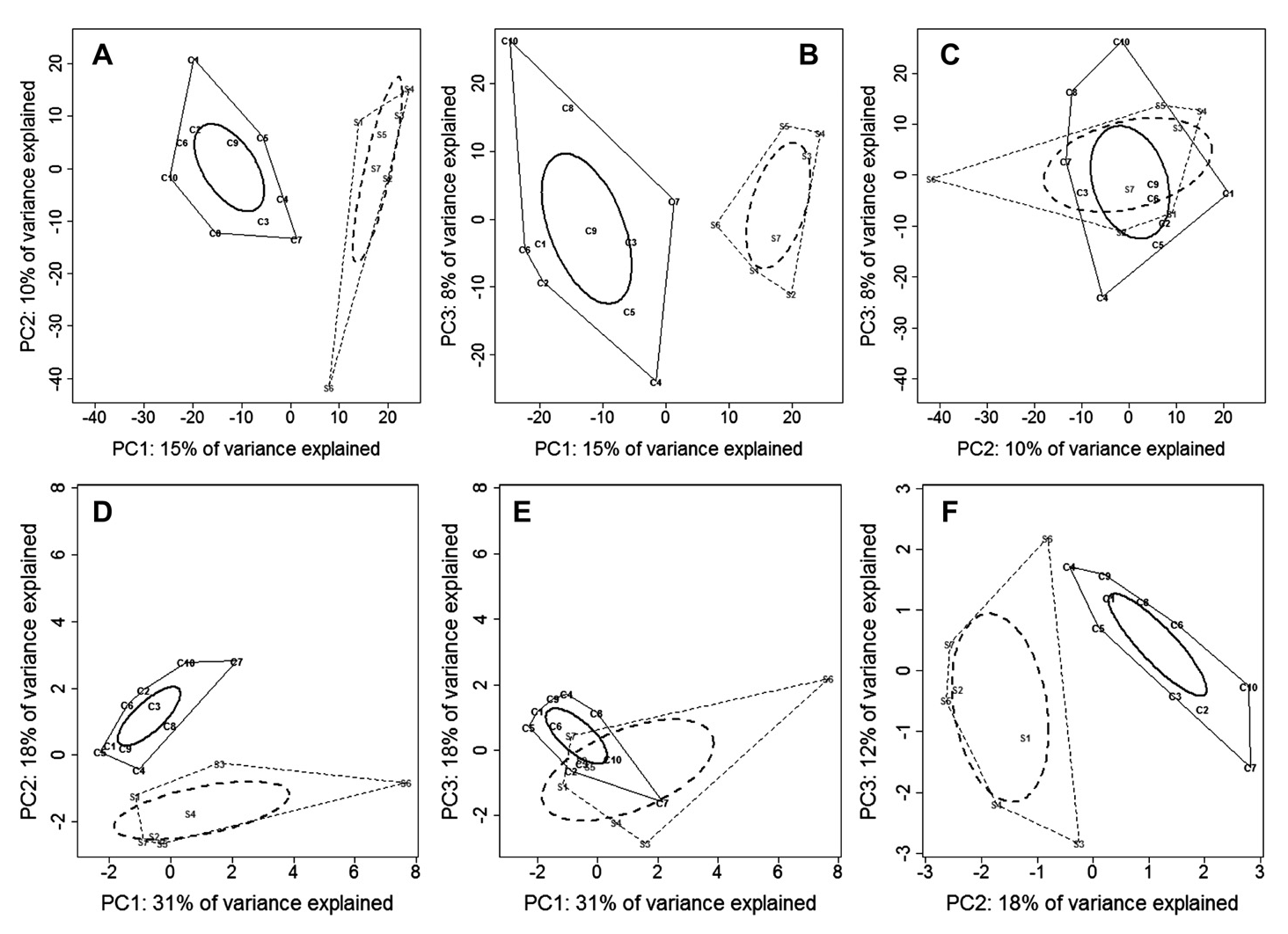
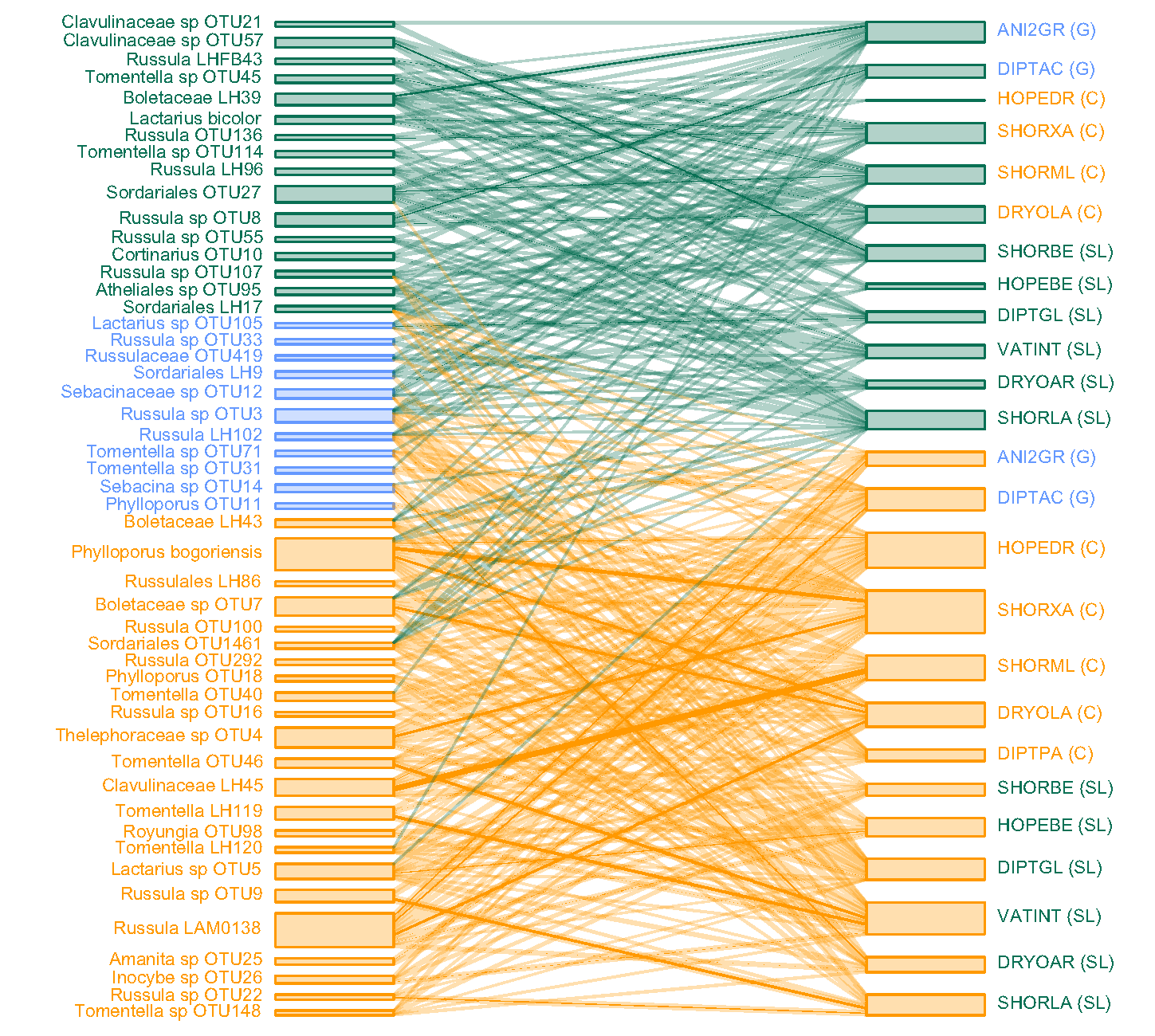
Plants interact with a diversity of microorganisms, and there is often concordance in their community structures. Because most community-level studies are observational, it is unclear if such concordance arises because of host specificity, in which microorganisms or plants limit each other’s occurrence. Using a reciprocal transplant experiment, we tested the hypothesis that host specificity between trees and ectomycorrhizal fungi determines patterns of tree and fungal soil specialisation. Seedlings of 13 dipterocarp species with contrasting soil specialisations were seeded into plots crossing soil type and canopy openness. Ectomycorrhizal colonists were identified by DNA sequencing. After 2.5 years, we found no evidence of host specificity. Rather, soil environment was the primary determinant of ectomycorrhizal diversity and composition on seedlings. Despite their close symbiosis, our results show that ectomycorrhizal fungi and tree communities in this Bornean rain forest assemble independently of host-specific interactions, raising questions about how mutualism shapes the realised niche (Peay et al. 2015), as well as how dominance by ectomycorrhizal tree species such as the Dipterocarpaceae arises (Fukami et al. 2017).
Despite the lack of host-specificity, mycorrhizal interactions, along with lithological variation, contribute to structuring the vast tree diversity in this Bornean rain forest. The proportion of biomass of ectomycorrhiza-hosting trees increased with declining soil fertility, with associated variation in functional traits. Our findings support a new framework integrating the mycorrhizal-associated nutrient economy and resource economic spectrum (Weemstra et al. 2020).
Plant-rhizobiome interactions in crop and grass species
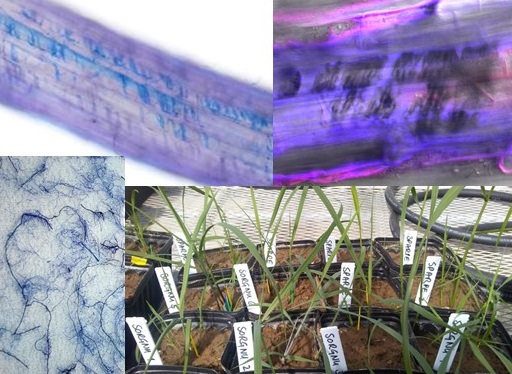
There is growing evidence that plants strongly influence the microbial community near their roots, also known as the rhizobiome. The ability to favor the growth of more beneficial microorganisms that enhance nutrient availability or uptake for plants or increase their resistance to soil pathogens should be selected for, but little is known about the mechanisms involved. The Russo lab is part of a multi-PI project (crri.unl.edu/about-crri) investigating such plant-microbe rhizosphere interactions in natural grassland and agricultural systems. The project uses an integrative approach to identify how microbial community structure and function influence plant phenotypes using stable-isotope probing, metagenomics, and proteomics.
Former PhD students Amanda Quattrone and Yuguo Yang studied how the interactions between plant roots, soil, and soil microorganisms affect plant productivity, root function, and drought resistance. Read more about some of their exciting work in these papers:
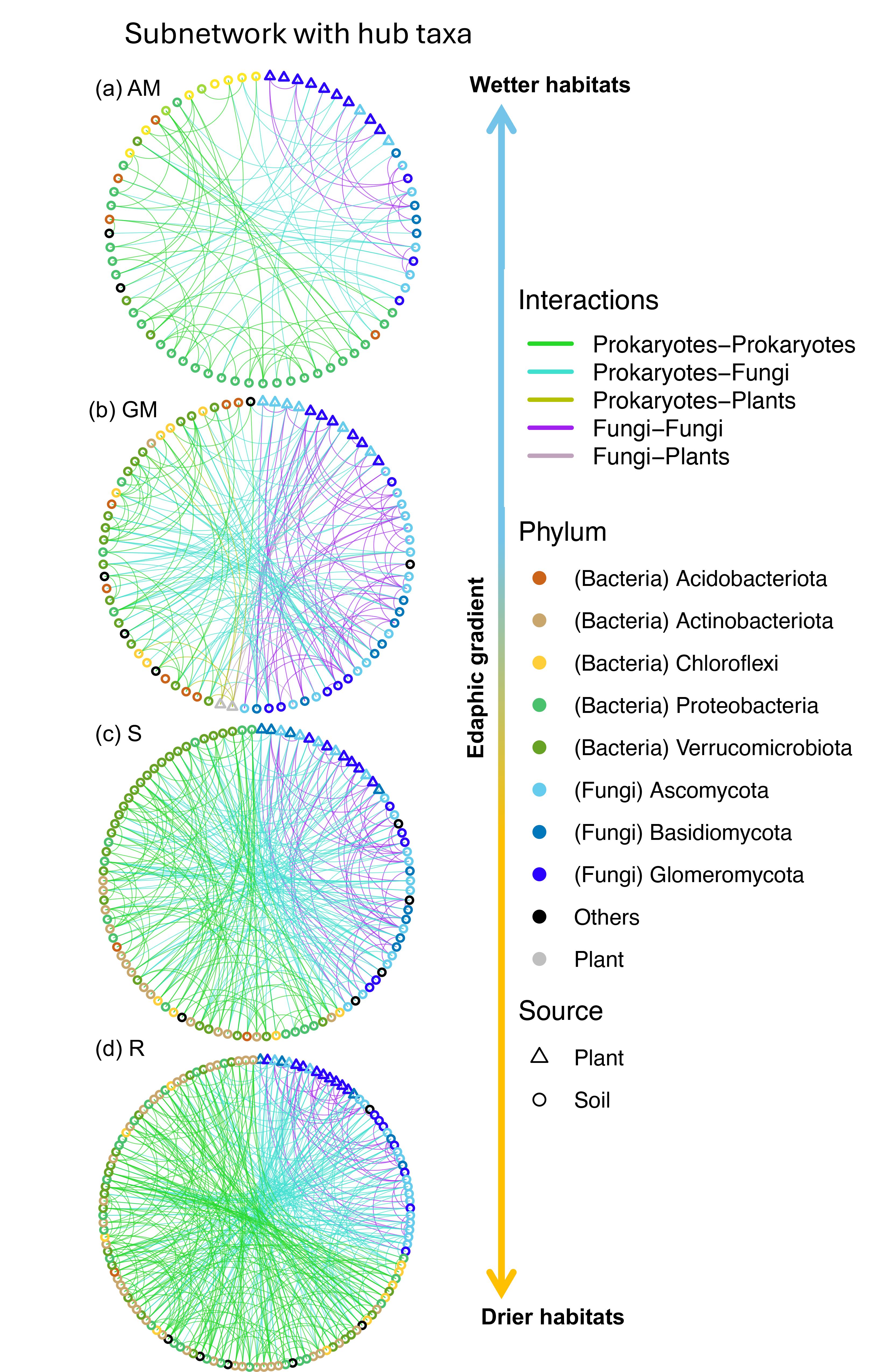
- Yang et al. 2025 Distinct roles of bacteria and fungi in belowground networks and predicting plant diversity and productivity in a North American grassland. FEMS Microbiology Ecology, accepted.
- Yang, Y. and S.E. Russo. 2024. Trade-offs in rooting strategy dimensions along an edaphic gradient in a grassland ecosystem. Functional Ecology 38:792–807 doi.org/10.1111/1365-2435.14514 ** Winner of 2024 Haldane Prize!
- Quattrone, A., Y. Yang, P. Yadav, K.A. Weber, S.E. Russo. 2023. Nutrient and microbiome-mediated plant-soil feedbacks in domesticated and wild Andropogoneae: Implications for agroecosystems. Microorganisms 11(12), 2978, doi.org/10.3390/microorganisms11122978
- Quattrone, A., M. Lopez-Guerrero, P. Yadav, M.A. Meier, K.A. Weber, S.E. Russo. 2023. Interactions between root hairs and the soil microbial community affect the growth of maize seedlings. Plant Cell and Environment doi.org/10.1111/pce.14755